Basic Molecular Biology Techniques understanding
Molecular Biology and Genetic Engineering — finechem
Molecular biology is built on a collection of essential techniques that allow scientists to study, analyze, and manipulate nucleic acids (DNA and RNA). These methods form the foundation for understanding genetic material and unlocking innovations in biotechnology and genetic engineering.
At finechem.org, we provide a comprehensive guide to core molecular biology techniques, including nucleic acid extraction and purification, quantification, amplification by PCR, electrophoresis, hybridization, sequencing, and more. Our content is designed to support students, researchers, and professionals in mastering these critical laboratory protocols.

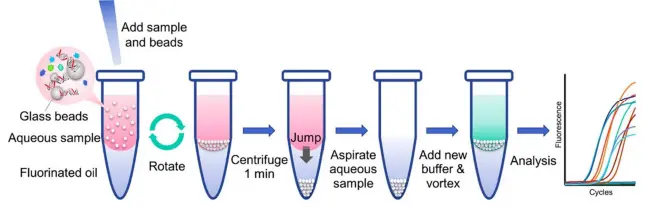
Nucleic Acid Extraction and Purification
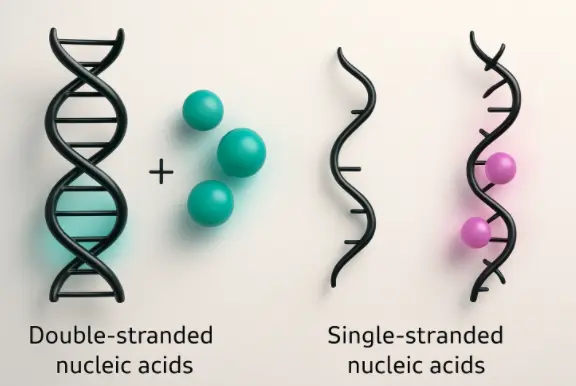
Nucleic acid extraction is the essential first step in any molecular study. This technique isolates DNA or RNA from cells or tissues by removing proteins, lipids, and other contaminants. Common methods include cell lysis, phenol-chloroform extraction, silica column purification, or magnetic bead separation.
Nucleic Acid Quantification
Quantification measures the concentration and purity of extracted nucleic acids. The most widely used methods are spectrophotometry (A260/280 ratio) and fluorescence-based assays using dyes such as PicoGreen or SYBR Green.
Nucleic Acid Labeling
Labeling involves attaching a detectable marker (radioactive, fluorescent, or enzymatic) to nucleic acids for subsequent detection.
Probe Labeling Techniques
- Nick translation: labeled nucleotides are incorporated during repair of single-strand nicks.
- Random priming (multi-random priming): synthesis of labeled probes using random primers.
- Synthetic oligonucleotide labeling: direct attachment of labels to synthetic sequences
Nucleic Acid Hybridization
Hybridization relies on the binding of complementary sequences.
Types of Hybridization
- Southern blot: detection of DNA fragments transferred onto a membrane.
- In situ hybridization: detection of sequences within cells or tissues.
- Chromosomal hybridization (FISH): detection of specific loci on metaphase chromosomes.
- Colony or plaque hybridization: detection directly on bacterial colonies or lysis plaques.
- Restriction Fragment Length Polymorphism (RFLP): analysis of genetic variation via restriction patterns
Discover more
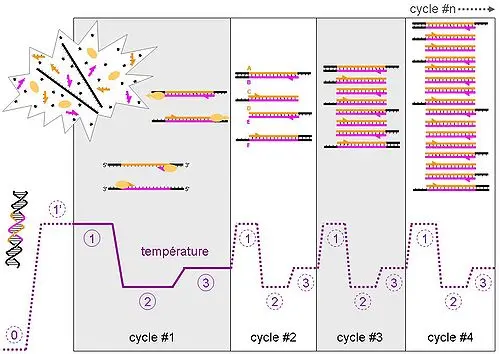
Sequencing
DNA sequencing is the process of determining the precise order of nucleotides (adenine, thymine, cytosine, and guanine) within a DNA molecule. This technique is fundamental for genetic analysis, mutation detection, molecular diagnostics, and many applications in biotechnology and genetic engineering.
Sanger Method (Chain Termination Sequencing)
The Sanger method, also known as chain termination sequencing, is based on the incorporation of labeled dideoxynucleotides (ddNTPs) during DNA synthesis. These modified nucleotides lack a 3'-OH group, preventing further extension of the DNA chain. By using a mixture of normal deoxynucleotides and fluorescently or radioactively labeled ddNTPs, DNA fragments of varying lengths are produced.
After capillary electrophoresis or gel electrophoresis, the sequence is determined by analyzing the pattern of terminated fragments. This method remains a gold standard for high-accuracy sequencing of individual genes and smaller DNA regions.
⚗ Maxam-Gilbert Chemical Method
The Maxam-Gilbert method relies on chemical modification of DNA bases, followed by base-specific cleavage at the modified sites. Each reaction targets a specific nucleotide or nucleotide type (e.g. G, A+G, C, C+T).
The resulting fragments are separated by electrophoresis, and the sequence is read from the pattern of cleaved fragments on the gel. While historically important, this method is less commonly used today due to the complexity of handling hazardous chemicals and the rise of safer, high-throughput sequencing technologies.
Our latest content
Check out what's new in our company !
Our latest content
Check out what's new in our company !